PCO - Protein Contacts Ontology
Introduction
Recent benchmark studies revealed that the accuracy and reliability of contact-site predictions delivered by state-of-the-art methods are low [Monastyrskyy et al., 2011]. They are insufficient for contact-based protein reconstruction. The reason for that may be the fact that most methods, with only a few exceptions [Walsh et al., 2009], treat all contacts as equal, based on a simple geometrical definition, despite that fact that only some of them interact and influence the overall protein structure. Identification of contact sub-types would allow to decompose the problem into smaller problems of predicting certain types of contacts, which might improve the accuracy of predictions. Also, better annotation of available structural data might contribute to our better understanding of the nature of protein contacts. To perform these tasks, a standardized formally defined vocabulary would be of great value. Although there are a number of ontologies used to describe structural features of proteins, like PFO (Protein Feature Ontology) [Reeves et al., 2008] or PLIO (Protein-Ligand Interactions Ontology) [Ivchenko et al., 2011], none of them was designed to model protein residue-residue contacts. PCO is the first ontology that allows to process protein data in terms of protein contact site descriptions.
The Aim
The aim of the ontology is to provide a formal representation of the agreed, existing knowledge in the domain of protein contacts, which allows unambiguous data annotation.
Ontology design
The ontology was developed in Protege editor (http://protege.stanford.edu) using Ontology Web Language 2 (OWL 2). Reasoners implemented in Protege were used to verify whether the ontology is consistent and coherent. In Figure A and Figure B the most generic terms of PCO hierarchy are depicted.
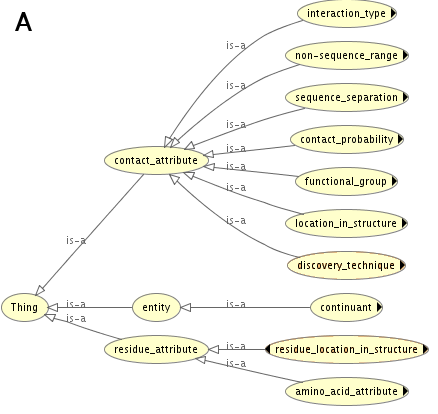
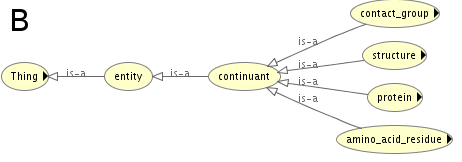
At this level the ontology has three distinct classes: i) 'contact_attribute', ii) 'entity', iii) 'residue_attribute'. The part of the ontology related to 'contact_attribute' defines attributes/properties that can be used to describe protein contact sites e.g. the type of observed physico-chemical interaction ('interaction_type'), the distance between amino acids in contact defined as their separation in the protein sequence ('sequence_separation') or localization of the contact within the protein ('location_in_structure'). The part of PCO related to 'residue_attribute' includes terms that allow to describe amino acid residues. The two specified aspects are: i) location and ii) physico-chemical properties resulting from the amino acid side chain. Finally, the terms grouped under 'entity' (Fig. B), are used to model objects such as protein structural regions, amino acid residues or contact sites. Following the guidelines provided by OBO consortium (Open Biomedical Ontologies Consortium) [Smith et al., 2007], fragments of other ontologies were reused, where possible. For instance the description of protein structural regions within PCO is mainly based on terms imported from the Sequence Ontology (SO) [Eilbeck et al., 2005]. The class hierarchy of PCO is based on two relations: 'is_a_subclass_of' and 'part_of'. However, other relation types were also defined for instance, 'has_interaction_between_residues', which is a relation between a contact site and a term that specifies the type of interaction between amino acids in contact. The formal description of contact sites allows to classify them with the use of Description Logic (DL). Apart from providing a framework for classifying contacts, the aim of the ontology was to enable integration of data from several sources. Therefore, in the the ontology there are classes for representing the fact that an instance of a particular residue-residue contact is present in multiple model-structures.
Download
The current version of PCO can be downloaded
here. The software for conducting protein annotation with ontology terms will be made available soon.
Contact

